Scanning through the rows and rows of preserved fish housed in the UW Fish Collection, it’s easy to get lost trying to figure out what each fish is, especially to the untrained eye. Fish identification is a necessary step when preserving specimens from the wild, which deliver key insights for researchers delving into the untold secrets of fish. What fish is it? Male or female? What age? Where was it collected from? These are just some questions answered before they’re put in jars to preserve for the future.
For Liam Aston, an undergraduate in his final year at SAFS, his capstone research involves Cottus, a group of freshwater sculpin found throughout the Northern Hemisphere. Although sculpin are found in both marine and freshwater environments, Liam is specifically focusing on the clade of Cottus found in Washington and two freshwater sculpins: Cottus gulosus (inland riffle sculpin) and Cottus perplexus (Reticulate sculpin). “These species are often confused in identification due to the overlap in identifying characters, leaving it to be decided by a coinflip most times,” Liam shared. “Determining which dichotomous keys – also known as identification tools – lack characters will further help identify sculpin in the future.”

Some of the characters found in keys used to identify sculpin are standard length (snout to the hypural plate), depth of fins, height of the dorsal connection, and mouth width. Part of Liam’s analysis of keys will also help to determine at what step identification error happens and therefore improve the process as a result. “I have been taking measurements of all the sculpins that are on the phylogenetic tree – also known as the family tree – completed by Álvaro Cortés, Vertebrate Collections Manager at Oregon State University. Using these measurements in combination with the tree, I will be able to create models to determine if there are any characters separating the species,” Liam said.
At SAFS, the capstone research project is the culmination of the undergraduate experience, an exciting opportunity to put classroom learning into practice and allow students to make a lasting contribution to aquatic and fishery science. For Liam, he chose to work with the UW Fish Collection’s Curator of Fishes, Luke Tornabene. “I had been interested in the work of the Fish Collection since I took FISH 311 with Luke in sophomore year, so this was a great opportunity to finish off my SAFS degree,” Liam said.
- Cottus asper (UW 158706)
- Cottus perplexus (UW 156980)
- Cottus gulosus (UW 159961)
When asked what his favorite part of his capstone research has been so far, Liam had a couple of things to share: “Going up to Friday Harbor Labs to use their modern CT scanner was pretty awesome because, for one, you get to visit the San Juan Islands, and two, using a CT scanner to scan a fish is cool.” A modern CT scanner takes roughly 30 minutes to scan a fish, when in the past, it would take 4-5 hours per fish! “It’s also been fun to learn from Katherine Maslenikov (Collections Manager) and Luke to gain a better understanding of taxonomy and the Fish Collection as a whole,” Liam added. Taxonomy is the scientific study of classifying, describing, and naming organisms.
If you’ve paid close attention to CT images or photos of fish specimens, they’re usually lying on their right sides, with their left sides featured in the image. Ever wondered why? This is because the standard in museum specimens is to cut material or take genetic samples from the right side of a fish, leaving the left side intact. So when you see the photos or scans of fish, they’re usually facing all the same way!

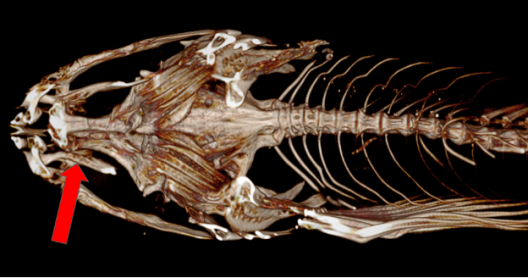
CT scanning is a very important tool when it comes to fish specimens. “One key identifying characteristic of freshwater sculpin is the internal presence of palatine teeth. The standard method to determine the presence of palatine teeth requires the jaw of the fish to be pried open, which can damage the specimen and still leaves uncertainty in the determination of palatine teeth presence,” Liam said. “Using the CT Scanner maintains the fish collection specimens held by the University of Washington and Oregon State University, and allows for the determination of palatine teeth to be certain. For the project I’m working on, many external counts and measurements need to be taken, but existing damage on collection specimens prevents accurate data from being collected. The CT Scanner circumnavigates the issue allowing accurate counts and measurements to be taken, plus these models can also show internal features that wouldn’t have been recognized just visually looking at fish specimens.”
- A side-on view of a Cottus gulosus via a CT Scan.
- Highlighting palatine teeth, this CT scan of Cottus gulosus is oriented looking up from the bottom.
One of the ways in which undergraduates conduct research for their capstone project is by working on research questions posed by faculty members. In Liam’s case, this happened to be freshwater sculpin. “I was most interested in freshwater fish, but I also wanted to do something involving speciation and phylogenetics. I wasn’t expecting to be separating species using morphometrics (for example, shape, form or size), but it has definitely grown my interest in taxonomy as a result,” Liam said.
As a culminating requirement of the SAFS degree, a key aim of a capstone is to put into practice the teaching and learning occurring over an undergraduate’s journey through SAFS classes. “A number of the classes I’ve taken while at SAFS have helped me with my research. FISH 311 introduced me to taxonomy and gave me a really strong understanding of phylogenetic trees and actually working through taxonomic keys, which has been a big part of this research to understand where the differences in species identification comes from,” Liam said. “FISH 290 helped me learn to read and understand scientific literature which has proved useful when going through literature for this project.”
Interested in other SAFS undergraduate research stories?